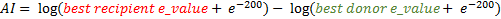FAQ
Frequently Asked Questions
How to calculate an Alien Index (AI), a HGT index and an AHS score
Alienness takes as input the result of a BLASTp search of a whole set of predicted proteins of interest (e.g. from a whole genome or a transcriptome) against the NCBI’s non-redundant (nr) library or any protein library available at NCBI.
The blast result of each query is read from the best blast hit to the last significant hit. Thus, the program records a couple of values composed of the best hit assigned to the taxonomic donor group called best donor e-value/score and the best hit assigned to the recipient taxonomic group called best recipient e-value/score.
Fields: query acc.ver, subject acc.ver, % identity, alignment length, mismatches, gap opens, q. start, q. end, s. start, s. end, evalue, bit score, taxid
Minc3s00019g01246 WP_028034051.1 50.6 338 163 3 1 338 1 334 1.4e-96 363.2 935840
Minc3s00019g01246 WP_011579798.1 51.2 334 159 3 5 338 5 334 1.6e-95 359.8 499972
Minc3s00019g01246 WP_159588070.1 49.5 333 164 3 6 338 6 334 2.5e-93 352.4 2681485
Minc3s00019g01246 KFB10895.1 50.0 332 162 3 8 339 8 335 2.8e-92 349.0 472175
Minc3s00019g01246 WP_051913977.1 50.0 332 162 3 8 339 10 337 2.8e-92 349.0 472175
...
Minc3s00019g01246 RCL30100.1 41.3 341 183 8 8 334 33 370 4.8e-60 241.9 2026799
Minc3s00019g01246 WP_088076312.1 42.2 344 178 7 1 330 1 337 4.8e-60 241.9 186817
Minc3s00019g01246 XP_009065820.1 42.1 340 181 10 1 331 1 333 4.8e-60 241.9 225164
Minc3s00019g01246 PYT47325.1 41.9 344 182 9 2 330 30 370 4.8e-60 241.9 1978231
Minc3s00019g01246 PCJ07871.1 39.6 338 197 6 1 338 1 331 4.8e-60 241.9 1904441
Minc3s00019g01246 WP_121651289.1 40.7 322 181 7 8 326 10 324 4.8e-60 241.9 1176649
...➮ The alien index is computed with the following formula :
In our example, the couple of best e-value is ( best recipient e-value : 1.4e-96 / best donor e-value : 4.8e-60 ) giving an AI equal to 84.13
Parameters Description AI Alien Index is a metric that allows to characterize the potential horizontal genes transfer best recipient e-value best BLAST E-value for the recipient taxon best donor e-value best BLAST E-value for the donor taxon
When either no donnor or no recipient significant BLAST hit is found, a penalty e-value of 1 is automatically assigned as the best donor or recipient e-value, respectively.
Hence, e-values of the best recipient and donor hits vary between 0 and 1 and, consequently AI scores vary between -460.5 and 460.5.
An AI>0 indicates a better hit to a donor species than to a recipient species and possible acquisition via HGT.
To know more, this method is defined in :
➮ The HGT index is computed with the following formula :
In our example, the couple of best score is ( best recipient score : 241.9 / best donor score : 363.2 ) giving a HGTindex equal to 121.30
Parameters Description HGT index HGT index is a metric that allows to characterize the potential horizontal genes transfer best recipient score best BLAST bitscore for the recipient taxon best donor score best BLAST bitscore for the donor taxon
➮ The AHS is computed with the following formula :
We developed a new metric called Aggregate Hit Support (AHS). We first normalise each bitscore Eq (1). We then sum all the normalised bitscores of the Donor hits and all the normalised bitscores of the Recipient (Ingroup) hits seperately and calculate the difference Eq (2). A positive AHS score suggests a potential HGT candidate.
What files and settings are expected
Input file
Alienness takes as input the BLAST or DIAMOND result of a proteome performed against a protein database.The input file for alienness tool must be compressed in .zip or .gz format.
For example, the blastp program is used and available on the NCBI ftp website in BLAST+ package.
Expected options for the command-line blastp :
-option value Description -outfmt 'X std staxid' X = 6 = Tabular
X = 7 = Tabular with comment lines-db nr BLAST database name
For a better coverage of the biodiversity, NCBI's nr library is recommanded but not necessary.
The protein library must have gi or accession numbers that exist in the NCBI database.-seg no The SEG program is used to mask or filter low complexity regions in amino acid queries -evalue 1e-3 Expect value (E) for saving hits
Alienness parameters
Alienness requires the user to define two taxonomic groups : the group of donor species and the group of recipient species. The value of the taxonomic node or NCBI TaxID entered in the field "taxonomic group of interest" (TOI) is used to define these two taxonomic sets, only 1 TaxID should be put. Thus, one will group all taxonomic nodes included in the provided taxonomic node (donor group), the other will contain all other nodes (recipient group).
For instance, if you are interested in HGT of non-metazoan origin to a metazoan species, please input 33208 (NCBI TaxID for Metazoa). If you are interested in HGT of non green plant origin to a green plant species, please input 33090 (NCBI TaxID for Viridiplantae).
This is valid for any TaxID and this information is necessary to retrieve the best ‘TaxID of recipient’ e-value and best ‘TaxID of candidate donor’ e-value for calculation of an Alien index.
Taxonomic group(s) to exclude
Alienness expects NCBI TaxIDs (one or several) for the taxonomic groups you want to ignore in the calculation of the Alien index. You must at least input the TaxID of the query species you used to produce the BLAST result. Anything that will be included in the entered taxonomic node will be excluded. Note that you can input several TaxIDs separated by comma in this field if you want to ignore several non-overlapping taxonomic groups. This is useful if there is no monophyletic group in the NCBI taxonomy corresponding to the ensemble of species you want to ignore.
Taxonomic group(s) used to classify potential donors
By default, the taxonomic groups found are categorized as Archaea, Bacteria, Viruses, Eukaryota, Eukaryota@Fungi, Eukaryota@Metazoa, Eukaryota@Stramenopiles and Eukaryota@Viridiplantae. ‘Other’ and ‘Unclassified’ groups are ignored as they cannot be assigned to a species. If left blank, the best hits are classified in these main categories. If you want to further classify the best hits in other categories any additional NCBI TaxID can be entered (e.g: Chorophyta, Streptophyta).
In summary ...
Alienness expects NCBI TaxIDs.
Fields Description Taxonomic group of interest TaxID, only one Taxonomic group(s) to exclude TaxID, one or several separated by comma Taxonomic group(s) to used to
classify potential donorsTaxID, optionnal, none to several separated by comma
Results description
The result of Alienness tool is a compressed directory to download, named Alienness_alienness-job-number.zip.Uncompressed directory contains :
Alienness_2017080515355122254/
├── Minc3_Metazoa_egp_Tylenchomorpha_alienness_AI_CALCULATION.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_alienness_FEATURES.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_alienness_INDEX.html
├── Minc3_Metazoa_egp_Tylenchomorpha_alienness_KRONA.html
├── Minc3_Metazoa_egp_Tylenchomorpha_alienness_SUMMARY.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_main.csv
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_queries_1_likely_hgt.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_queries_2_possible_hgt.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_queries_3_likely_contamination.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_taxonomy_0_all_hgt.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_taxonomy_1_likely_hgt.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_taxonomy_2_possible_hgt.xls
├── Minc3_Metazoa_egp_Tylenchomorpha_stat_taxonomy_3_likely_contamination.xls
├── html/
└── src/
The result files contain the following information :
Results files description Project_name_alienness_AI_CALCULATION.xls (*1) table presenting AI, HGT score and AHS score values and taxonomic information
for all the proteins that returned an AI valueProject_name_alienness_FEATURES.xls (*2) AI features file necessary to use AvP Project_name_alienness_INDEX.html an html index file that allows visually exploring the BLAST results with a color code Project_name_alienness_KRONA.html Krona charts are created to explore the best donors detected in your dataset Project_name_alienness_SUMMARY.txt log file providing information on execution time, parameters selected by the user, ... Project_name_stat_main.xls (*3) number of queries classified in each category (likely hgt; possible hgt; likely
contamination; not hgt)Project_name_stat_queries_1_likely_hgt.xls (*4) list of queries with a AI > 15 Project_name_stat_queries_2_possible_hgt.xls (*4) list of queries with a AI between 0 and 15 Project_name_stat_queries_3_likely_contamination.xls (*4) list of queries with a AI > 15 and a percentage of identity > 70 Project_name_stat_taxonomy_0_all_hgt.xls (*5) statistics on the taxonomic distribution (species and kingdoms) of candidate donors
for all categoriesProject_name_stat_taxonomy_1_likely_hgt.xls (*6) statistics on the taxonomic distribution (species and kingdoms) of candidate donors
for the likely HGT categoryProject_name_stat_taxonomy_2_possible_hgt.xls (*6) statistics on the taxonomic distribution (species and kingdoms) of candidate donors
for the possible HGT categoryProject_name_stat_taxonomy_3_likely_contamination.xls (*6) statistics on the taxonomic distribution (species and kingdoms) of candidate donors
for the possible contamination category
We tested the accuracy of Alienness on the genomes of two plant-parasitic nematodes, for which phylogenetically supported HGT of a whole series of genes involved in plant parasitism had been previously identified [Danchin et al. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656] [Haegeman et al. Mol. Plant Microbe Interact. 2011]. We found that all phylogenetically supported cases could be retrieved by Alienness with an AI > 9 and that this AI threshold corresponded to a low rate of putative false positives. To focus on candidates that are likely to produce phylogenetic trees supporting HGT, and minimizing the rate of false positives, we recommend an AI > 15.
Three categories are defined :
* likely_hgt : AI > 0 and <70% identity to putative donors
* possible_hgt : 0 < AI < 15
* likely_contamination : AI > 0 and >70% identity to putative donors
Below, you can see the generic description of files marked by (*1)
Column num Column title Description 1 AI Alien Index calculation 2 HGTindex HGT index calculation 3 AHS Aggregate Hit Support calculation 4 query name the query description 5 category Four ordered categories from most likely hgt to rejected hgt
1:likely_hgt > 2:possible_hgt > 3:likely_contamination > 4:not_hgt6 query hits number the number of hits returned by the protein in consideration 7 acc_recipient best accession number for the user-defined taxonomic group of interest (recipient group, e.g Metazoa
or Stramenopiles)8 evalue_recipient best e-value for the user-defined taxonomic group of interest (recipient group, e.g Metazoa
or Stramenopiles)9 bitscore_recipient best e-value for the user-defined taxonomic group of interest (recipient group, e.g Metazoa
or Stramenopiles)10 acc_donor best accession number for the potential donor 11 evalue_donor best e-value for the potential donor 12 bitscore_donor best e-value for the potential donor 13 best closer to the best hit (donor hit or recipient hit) 14 best_hit_acc accession number of the closer to the best hit 15 best hit prct ident the percent identity between the query sequence and the best hit 16 best hit org full name full species name of the best hit 17 best hit taxo group abbreviated taxonomic classification of the best hit 18 best hit taxid NCBI taxID of the best hit 19 best hit lineage full taxonomic lineage for the best hit
Description of features file (*2)
Column num Column title Description 1 query_name the query description 2 donor donor information separated by ":"
info1:info2:info3:info4:info5 (*)3 recipient recipient information separated by ":"
info1:info2:info3:info4:info5 (*)4 AI Alien Index calculation 5 HGTindex HGT index calculation 6 AHS Aggregate Hit Support calculation 7 query hits number the number of hits returned by the protein in consideration
In addition, all the files named with a _stat suffix
(*3) main statistics
(*4) are built on a same twelve-column template and provide basic statistics on the candidate donors (or contaminant)
Column num Column title Description 1 HGT classification classification into three categories : likely_hgt > possible_hgt > likely_contamination 2 nb occurrence of each taxonomy
(*5) occurence of the best donors by taxonomy for each HGT category
Column num Column title Description 1 ai Alien Index calculation 2 hgtindex HGT Index calculation 3 AHS Aggregate Hit Support calculation 4 query the query description 5 best_donor_acc / best_toi_acc accessions of the donor/recipient couple 6 best_donor_pident the percent identity between the query sequence and the best hit donor 7 best_donor_orgname full species name of the best hit donor 8 best_donor_taxonomy abbreviated taxonomic classification of the best hit donor 9 nb_hits_supporting_taxo number of hits supporting the donor taxonomic group 10 nb_hits_between_donor_and_possible_toi number of hits found between the donor and the possible taxon of interest (recipient) 11 nb_total_hits total number of hits found 12 nb_unknown_acc (dr) number of unknown accessions found between the donor and the possible taxon of interest (recipient) 13 nb_excluded_acc (dr) number of excluded accessions found between the donor and the possible taxon of interest (recipient)
(*5) occurence of the best donors by orgname for each HGT category
Column num Column title Description 1 best donor by taxonomy taxonomic group 2 likely_hgt occurrence 3 possible_hgt occurrence 4 likely_contamination occurrence
(*6) are built on a same tthree-column template and provide basic statistics on the candidate donors (or contaminant)
Column num Column title Description 1 best donor by orgname organism name sorted by taxonomic group 2 likely_hgt occurrence 3 possible_hgt occurrence 4 likely_contamination occurrence
(*6) are built on a same tthree-column template and provide basic statistics on the candidate donors (or contaminant)
Column num Column title Description 1 HGT classification classification into three categories : likely_hgt > possible_hgt > likely_contamination 2 best donor by taxonomy taxonomic group 3 nb occurrence
Column num Column title Description 1 HGT classification classification into three categories : likely_hgt > possible_hgt > likely_contamination 2 best donor by orgname organism name 3 nb occurrence